About
Toxicogenomics is based on the vast improvements of genomic tools over the last decades. Those tools make it possible to analyse genomic expression modifications on a large scale that can be linked to effects of substances within the organism. The word «omics» thereby means that the analysis is done for the whole amount of the structure to be analysed within the cell like the whole set of all expressed RNAs in one go rather than only one single specific RNA, for instance. One goal of toxicogenomics is to predict toxicity of compounds by genomic modifications that are induced in cells. In silico approaches rely on already existing data that can be used and since many relations between genomic variability and compound remain unclear, further experimentation has to be done to be able to make accurate in silico predictions. Nevertheless, the efforts and progress in this field are signficant and authorities have begun to regulate risk assessment for the field of toxicogenomics.
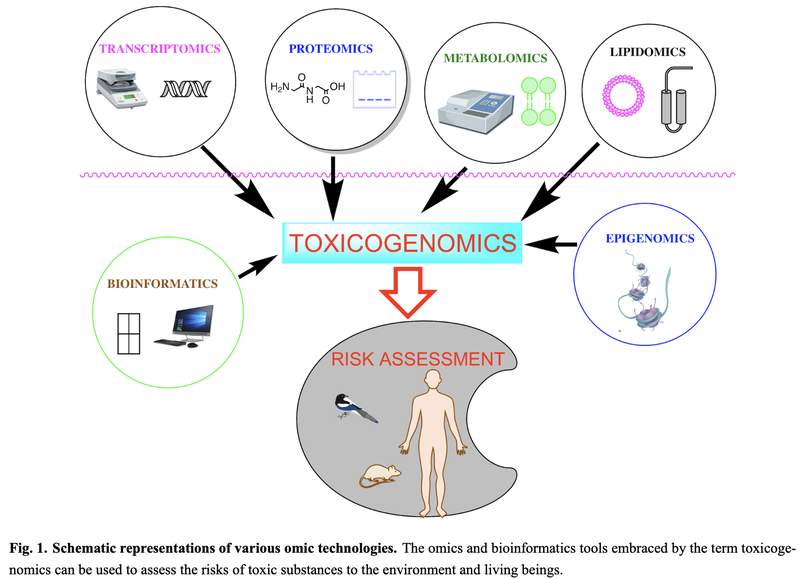
[2]
The technologies which are used for toxicogenomic analysis have improved during the last years and include transcriptomics, proteomics, metabolomics, lipidomics, epigenomics and bioinformatics [2]. Thereby,
Transcriptomics allows to measure genome wide gene expression by using RT/qPCR-, DNA microarray- or total RNA sequencing techniques.
Proteomics is a tool with which we can measure the presence of proteins and their modifications. This can be done by gel-electrophoresis on a small scale and more precisely by Mass Spectroscopy (MS) for a large amount of datasets. By the improvement of sample preparation techniques, MS instrumentation and data access at databases, it is possible to quantify more than 1000 different proteins from one single cell.
Metabolomics and lipidomics provides the possibility to measure the amount and structure of metabolites and lipids by using Liquid- and Gas Chromatography followed by MS techniques and NMR.
Epigenomics describes the analysis of epigenetic modifications such as histone methylation for instance. Such modifications can be measured by techniques such as the genome-wide bisulfite sequencing.
Bioinformatics is important in toxicological risk assessment since single omics techniques are not enough to characterise compounds in concern of toxic effects. Multi omic techniques have to be combined and analysed where not only proper computational capacity is required but also to valuable datasets which have to be accessible in databases. Such databases include The Gene Ontology Database, The Kyoto Encyclopedia of Genes and Genomes (KEGG), The Genomics of Drug Sensitivity in Cancer Database (GDSC), the Comparative Toxicogenomics Database (CTD), The Expression Atlas and the SWISS-Prot protein-specific database.
References
[1] Application of Toxicogenomic Technologies to Predictive Toxicology and Risk Assessment by The National Research Council in The National Academies Press, 2007.
[2] Perspective on the Use of Toxicogenomics to Assess Environmental Risk by José Portugal, Sylvia Mansilla and Benjamin Pina in Frontiers in Bioscience Landmark, 27(10): 294, 2022
